List of Publications
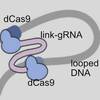
Re-engineered guide RNA enables DNA loops and contacts modulating repression in E. coli
Can you do "DNA origami" inside a cell? Maybe, if you learn how to fold double-stranded DNA in cellulo. Yunshi developed this idea using two dCas proteins linked with a common guide RNA that target two sites on DNA. It can fold DNA loops in bacteria and modulate gene expression.

A DNA robotic switch with regulated autonomous display of cytotoxic ligand nanopatterns
Following our work on TRAIL peptide clustering of Death Receptors, we noticed that the pattern we discovered was highly cytotoxic, so using it for cancer therapy would indiscriminately harm cells and tissue. So Yang figured he would hide the peptides in a barrel-like origami, and display the cytotoxic pattern only at low pH using triplex-DNA formation. This became a logic-gated nanorobot, killing cancer cells only in solid tumors!
An error correction strategy for image reconstruction by DNA sequencing microscopy
Continuing with our work of making images from neighbour-neighbour networks deduced from sequencing data, Alexander finds some very clever ways to spot false connections between nodes that probably were never neighbours in the first place. Our second publication in the new field of DNA microscopy.

Soluble and multivalent Jag1 DNA origami nanopatterns activate Notch without pulling force.
Using DNA origami to arrange molecules on the nanoscale is a powerful way to obtain new biological knowledge. Here Ioanna uses nanostructures decorated with the Notch-ligand Jag1 and discovers that Notch receptors can be activated by these structures without applying a pulling force on the receptor. Could this be used to make new Notch agonists for therapy or stem cell engineering?
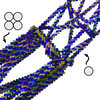
Computer-Aided Design of A-Trail Routed Wireframe DNA Nanostructures with Square Lattice Edges
Polygonal origami using A-trail routing of the scaffold is the most efficient way to make a DNA origami mesh as you can use one helix per edge. Sometimes however, you want to reinforce some of these single helices, to increase rigidity at that place. Marco has made this process very simple by automating conversion of some edges from 1 to 4 helices.
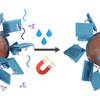
Solid Phase Synthesis of DNA Nanostructures in Heavy Liquid
What if DNA origami could be folded on solid supports? That would simplify purification and subsequent addition of functional molecules. In this work Ioanna realizes such an application using a heavy dense liquid to keep magnetic beads afloat during the whole folding process!

Stochastic modeling of antibody binding predicts programmable migration on antigen patterns
Can antibodies (Abs) walk around on patterns of antigens, and can we pattern antigens to drive the walking towards specific locations? We think so! Based on our previous data on Ab binding dynamics (see Nat. Nanotech. paper from 2019), Ian created a stochastic model to be able to predict binding behavior to complex patterns.

DNA Origami Penetration in Cell Spheroid Tissue Models is Enhanced by Wireframe Design
In this work we discover considerable differences between DNA origami interactions with cells depending on the internal structure of the DNA nanostructures. Wireframe, mesh-like, origami gets uptaken by cells in a slower, more controlled manner than close-packed origami. In particular the wireframe structures can penetrate deeper into cell spheroid tissue models and appear to interact less with scavenge receptors than their close-packed counterparts.
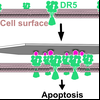
Clustering of Death Receptor for Apoptosis Using Nanoscale Patterns of Peptides
Nanoscale clustering of receptors at the cell surface is an important communication pathway for multicellular organism. Until the development of DNA nanotechnology, probing these clustering events precisely was very difficult. Here Yang used flat DNA-origami decorated with peptides that mimic the TRAIL-protein to learn how human cells react to these nanopattern signals.
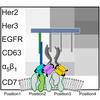
A DNA-nanoassembly-based approach to map membrane protein nanoenvironments
While there are many tools out there to look for protein pairs, there are very few tools to systematically investigate protein environments. In this collaboration Elena and Ana's lab developed a method based on barcode copying of binder-conjugate strands to get ensemble averaged measurements of nanoscale protein arrangements on cell membranes.
Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR
When Covid-19 struck we started thinking about ways to address bottlenecks in the qPCR testing with our neighboring lab of Björn Reinius. Reinius and PhD student Ioanna Smyrlaki in my lab worked hard in the spring to prove that actually just heating the patient sample and running extraction free RT-pPCR on it directly is more than enough to retain a highly sensitive test for SARS-CoV2 infection.

Evolutionary Refinement of DNA Nanostructures Using Coarse-Grained Molecular Dynamics Simulations
If we could simulate multiple designs and select the ones that perform best in simulations, modify them and simulate again and start over, we could potentially make DNA origamis with improved stability, stiffness and fit for purpose, without experimental iterations. Here, Erik shows the first steps in this direction.

A computational framework for DNA sequencing microscopy
Imagine tagging molecules inside a cell or tissue sample with random DNA barcodes. If we can make these DNA tags connect with each other enzymatically and then read all connected sequences, we could figure out the positions of all tags and their attached molecules relative to each other - image reconstruction by sequencing or DNA sequencing microscopy!

Binding to nanopatterned antigens is dominated by the spatial tolerance of antibodies
Antibodies can stretch and flex to bind with their Fab-arms to two antigens. But how well can they actually do it? Here we show for the first time how binding varies with distance between antigens and find some important clues that could potentially be used for vaccine design.

Solution-Controlled Conformational Switching of an Anchored Wireframe DNA Nanostructure
Thin DNA origami sheets can be quite flexible. Here, Ian attached sheets of origami to a surface and was able to controllably modulate the conformation of the sheet. By changing buffer the sheets would open and close like flowers. Optical superresolution imaging using DNA paint was used to monitor the process, and cryoEM revealed the inner structure of the meshes.

Effects of Design Choices on the Stiffness of Wireframe DNA Origami Structures
In macroscopic construction work, trusses are often used to increase rigidity while keeping the amount of building material used at a minimum. Here, Erik did the same thing but on the nanoscale. We investigated numerous was to make rigid beams using triangulated DNA structures and did molecular dynamics simulations in oxDNA to guide us on the way.
BtsCI and BseGI display sequence preference in the nucleotides flanking the recognition sequence
Our synthetic biology approach to making oligonucleotides using biotechnological mass production (the MOSIC method) relies on restriction enzymes to work the way we think they do. Here Joao shows that these types of restriction enzymes are a bit picky when it comes to the sequence of the DNA they cut also outside of the recognition site. We have added more enzyme in the reactions, now we know why that is crucial.
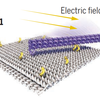
Remote control of nanoscale devices
Perspective about the work from Fritz Simmel's lab about an origami device that uses a platform and an arm that can be controlled by externally applied fields. A nice demonstration of the precision control of the molecular world made possible by DNA Nanotechnology.

Entirely enzymatic nanofabrication of DNA–protein conjugates
Many applications in synthetic biolog and bionanotechnology rely heavily on DNA-protein conjugates. DNA can be used to program reactions and structures and proteins can do actual work. Here, Giulio shows how one peculiar protein from Agrobacterium can be modified and harnessed to create DNA-protein conjugates on the fly entirely without synthetic chemistry. He also demonstrates some nice applications like probes and a novel transcriptional activator.

Measuring true localization accuracy in super resolution microscopy with DNA-origami nanostructures
In superresolution microscopy, like for example STORM and STED, how do one measure the actual resolution? In a lot of cases, one just looks at a single emitter and estimates the spatial spread of the signal. This is actually not a good way to do it. Here, Ferenc toghether with people in the Brismar lab show that origami is an excellent tool to characterize and compare these microscopy methods and enables a direct measure of how the signal spreads.

Computer-Aided Production of Scaffolded DNA Nanostructures from Flat Sheet Meshes
Building on our earlier work on polyhedral shapes (see below) we are now using a similar technique to render flat sheets in DNA. Interestingly, the original algorithm handled 3D objects much better than flat sheets using some mathematical tricks and lots of AFM imaging Erik and Melik were able to demonstrate how the technique can render some stunning flat sheet shapes that are larger (per base-pair) than the original flat sheets by Rothemund, and that fold without magnesium!

DNA rendering of polyhedral meshes at the nanoscale
Wouldn't it be nice to be able to just draw an nanostructure shape using conventional 3D graphics software? In this paper we show that this is now possible! Using a routing of the DNA scaffold that is derived from graph theory, we can automate the entire DNA-nanostructure design process and Erik used this to fold some pretty interesting shapes. Including a well known computer-graphics bunny...
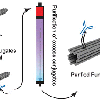
Purification of Functionalized DNA Origami Nanostructures
Using nanocalipers we can tinker with cell communication (see 'Spatial Control ...'-paper below). Now, how do you actually produce nanocalipers in the most efficient, purest way? In this paper Alan shows you how it is done for a wide range of purification methods using 3 different molecules that we like to put on our nanocalipers. And, introducing two new ways to purify DNA origami!
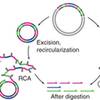
Rolling circle replication requires single-stranded DNA binding protein to avoid termination and production of double-stranded DNA
Rolling circle amplification (RCA), maybe not so rolling after all? We have found that Phi29 polymerase is not always following the right template when making new DNA in RCA. Sometimes the polymerase flips and starts to backtrack. This leads to an unexpected amount of double-stranded DNA where we initially expected to find only single-stranded DNA.

Spatial control of membrane receptor function using ligand nanocalipers
How do cells feel their environment? Can they sense nanoscale distribution of ligands at the surface of neighboring cells? Using DNA origami, we were able to show together with Ana’s lab that indeed it seems like cells do have some kind of blind-reading alphabet that they use to communicate.
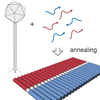
DNA Origami Structures Directly Assembled from Intact Bacteriophages
An origami design intended to fold the entire Lambda genome using 3000 oligonucleotides. We noticed that Lambda DNA that you buy is full of nicks and very unsuitable for use as an origami scaffold. So we made our own Lambda DNA from phage. And then Philipp just made phage and added the oligos directly to that...
Enzymatic production of 'monoclonal stoichiometric' single-stranded DNA oligonucleotides
Everyone needs DNA oligos. Well, maybe not everyone, but everyone who does some kind of biotechnical research at least. Solid-phase synthesis is great, but biological nanomachines (enzymes) kick-ass when it comes to producing long, sequence controlled polymers. Here, Cosimo and the lab show you how to make single-stranded DNA oligos enzymatically, and how this can be used in for example DNA nanotechnology. Check our 'software' section for a tool to calculate the pseudogenes!
A DNA Origami Delivery System for Cancer Therapy with Tunable Release Properties
DNA Origami can be used to deliver anti-cancer drugs! In fact, we found that by tuning the DNA twist density, we could create structures that really like to hold on to its Doxorubicin. The slow release allowed us to use less Dox than usual to get a therapeutic effect. More treatment with less is of course extra good when dealing with cardiotoxic drugs like Dox.

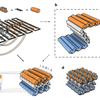
Self-assembly of three-dimensional prestressed tensegrity structures from DNA
By using DNA origami, Tim was able produce nanoscale tensegrity structures. The structures are composed of rigid rods and flexible bands. The rods were made from DNA origami bundles, and the bands were just single stranded DNA! It turns out that single stranded DNA makes perfect nanoscopic rubber bands. The tensegrity devices could be used to amplify small mechanical signals into large conformational changes: cutting one of the rubber bands will make the entire structure blow up.

Folding DNA Origami from a Double-Stranded Source of Scaffold
For DNA origami we need scaffolds, long single stranded DNA that can be folded with the help of short, synthetic DNA oligonucleotides. These long scaffold strands had up until this publication been limited to the genome of a single stranded DNA virus, the M13 virus. In this paper we show that using a few tricks you can actually use double stranded DNA and fold each of the two strands into separate structures! This will open the door to using a much broader variety of DNA samples for DNA origami.
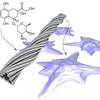
Self-assembly of DNA into nanoscale three-dimensional shapes
DNA origami, now in 3D! It turns out that Paul Rothemunds DNA origamis can be folded like real origamis into thick multi-layered object. We show off some pretty complex structures and try to explain the design process and experimental tricks in depth. Each of us contributed with a design that exemplifies different aspects of the technology. We had to do the design the hard way: writing scripts in python to generate the staple sequences. Fortunately for future users of this technology, Shawn's software caDNAno, nowdays makes the process a little less tedious.

Anisotropically Functionalized Nanoparticle Dimers
Building blocks for self-assembly need to have a minimal structural complexity, they can't look the same all around. They need to have an _up_ and a _down_, i.e. they must be anisotropic. Usually gold particles are pretty symmetric but in this paper we show that the trick to break the symmetry that we described in earlier papers is working in the lab.

DNA-Mediated Self-Assembly of Nanostructures - Theory and Experiments
This thesis summarizes the work Björn performed during the last years of his PhD at Mid Sweden University. Its an easy-to-read text with technical stuff mostly in the appended papers. Should be simple to follow even for those of you that are not familiar with nano self-assembly.
, p. (2007)

DNA Scaffolded Nanoparticle Structures
This paper describes some of our efforts to attach nanoparticles on DNA-origami. We reproduce Rothemunds original experiment and try to extend the concept by attaching proteins and metals to the origamis. Proteins are OK, attaching metal nanoparticles seems harder...

Programmable Self-Assembly - Unique Structures and Bond Uniqueness
If one wants to self-assemble a complex nanostructure, what are the basic requirements for the constituting building blocks? In this paper we try to look at the design level tradeoffs between making a few quite simple building blocks with few types of bonds, or a lot of building blocks with highly specific bonds. Surely the first alternative must be better?

Study of DNA coated nanoparticles as possible self-assembly building blocks
This paper describes our idea of how to make building blocks for algorithmic assembly from DNA-coated gold particles. If you coat a gold particle with DNA you will get a completely symmetric building block. Here we were one of the first to show a feasible way to break the symmetry and a path to build complex DNA-gold assemblies.

Novel in-situ Fabricated Josephson Junctions: Trilayer on a Substrate Slope
Back in the day when top-down fabrication was still a-la-mode, Bjorn put together a new approach for fabrication of superconducting Josephson-junctions. Thin films of YBaCuO were fabricated and processed in a cleanroom. Electronic measurements at 4 degrees Kelvin revealed that the junctions were working, sort of.
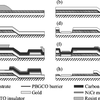
Submicron YBa2Cu3Ox ramp Josephson Junctions
Using electron beam lithography and state of the art technology for fabricating high temperature superconductors we managed to make functional Josephson junctions that were really small. Some interesting voltage - current characteristics were observed and discussed.